At work we were just migrated to o365 (‘M365’ as they call it), and :
* they’ve disabled forwarding (so can’t automatically forward my mail to e.g. a gmail account)
* they’ve disabled access for all email clients other than Outlook
This happened half a year ago to the students at Umeå, and here’s the solution Mats Wahlberg (Physics and Applied Mathematics) sent me:
1. Get evolution-ews
— on thunderbird you have the non-free (as in beer) options of OWL for exchange and ExQuilla
— DAVmail may also be a solution, but I haven’t managed to get it to work. It will act as a layer between your mail client (e.g. thunderbird) and the EWS server, allowing you to continue using e.g. thunderbird without having to install anything.
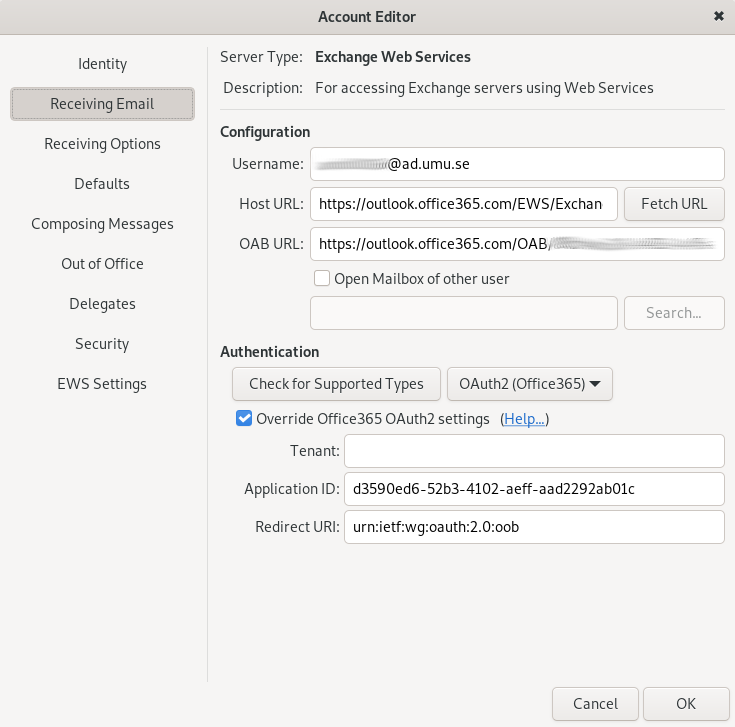
2. Create a ‘New Mail Account’:
Identity: do whatever is appropriate for your identity. Disable ‘Look up mail server …’
Receiving Email: Add the appropriate username (format: abcd0123@ad.umu.se) for connecting to the mail server.
Host URL: use https://outlook.office365.com/EWS/Exchange.asmx
Authentication type: OAUTH2 (Office 365)
Check ‘Override Office365 OAuth2 settings’
Tenant: leave empty
Application ID: d3590ed6-52b3-4102-aeff-aad2292ab01c
Redirect URI: urn:ietf:wg:oauth:2.0:oob
Then click on Fetch URL. This should populate the OAB URL field.
Done!

